About Human Gene and Protein Database (HGPD)
The entire human genome sequence has been determined by international project teams (1). In the post-genomic research era, one of the most essential subjects involves functional and structural analysis of gene products (proteins). To obtain full-length cDNA clones in hand is one of the key issues in such studies of functional genomics. Projects such as the Japanese FLJ project supported by NEDO (New Energy and Industrial Technology Development Organization) (2-4), the Kazusa long cDNA project supported by Chiba prefecture (a local government) (5, 6), the US Mammalian Gene Collection (MGC) program (7), German (8), Chinese (9) and other cDNA projects, have been implemented to isolate as many full-length cDNAs and as in a high quality as possible (For a review, see reference 10). To build the infrastructure to allow systematic and comprehensive expression of human proteins, not only the availability of full-length cDNA clones, but also a versatile system for making use of these clones is vital. The Gateway cloning system (Invitrogen, CA, USA) is based on such versatile expression vectors (11). We have therefore adopted this system and constructed human Gateway entry clones from full-length cDNAs (12). For conversion to Gateway entry clones, we first determined an open reading frame (ORF) region in each cDNA meeting the criteria (13,14). Those ORF regions were PCR-amplified utilizing selected resource cDNAs as templates. All the details of the construction and utilization of entry clones will be published elsewhere (12). Amino acid and nucleotide sequences of an ORF for each cDNA and sequence differences of Gateway entry clones from source cDNAs are presented in the "GW: Gateway Summary" window (Fig. 18). Utilizing those clones with a very efficient cell-free protein synthesis system featuring wheat germ (15,16), we have produced a large number of human proteins in vitro. Expressed proteins were detected in almost all cases (12). Proteins in both total and supernatant fractions are shown in the "PE: Protein Expression" window (Fig. 19). In addition, we have determined subcellular localizations of human proteins fused with the fluorescent protein in HeLa cells (17). The image data are shown in the "SL: Subcellular Localization" window (Fig. 20). These biological data are presented on the frame of cDNA clusters in the Human Gene and Protein Database (HGPD, http://www.HGPD.jp) (18). To build the basic frame of HGPD, sequences of FLJ full-length cDNAs and others deposited in public databases (Human ESTs, RefSeq, Ensembl, MGC, etc.) are assembled onto the genome sequences (NCBI Build 35 (UCSC hg17)).
In the NEDO full-length human cDNA sequencing project (FLJ-PJ), in addition to about 30,000 human full-length cDNAs (FLJ cDNAs) (2), about 1,430,000 5'- and 3'-end sequences (ESTs) of full-length cDNAs were deposited to DDBJ/GenBank/EMBL (3). These were obtained from cDNA libraries consisting of mRNAs for about 100 kinds of human tissues and cells constructed using the oligo-capping method. The majority of the insert cDNA sizes were over 2 kb and the full-length rate of 5'-end was more than 90% (19). By developing efficient search and evaluation systems for splicing variant (SV) cDNAs, more than10,000 important splicing variant cDNAs have been obtained. Then the FLJ Human cDNA Database (ver. 3.0) displaying those data has been constructed and will be up shortly (19). Only the number of SV cDNAs, which is 5,020 in total, is presented in the "others in d box" of the "C1: cDNA Summary 1" window in HGPD (Fig. 15). Most of those SV cDNAs have also been converted to Gateway entry clones.
*) The majority of analysis data for cDNA sequences in HGPD are shared with the FLJ Human cDNA Database (http://flj.hinv.jp/) constructed as a human cDNA sequence analysis database focusing on mRNA varieties caused by variations in transcription start site (TSS) and splicing.
What's New !
2011
| Date | Topics |
|---|---|
| 15 Sep |
|
Data Summary
HGPD entries
| Category | Numbers | |||
|---|---|---|---|---|
| Human cDNAs | FLJ cDNAs | Entirely sequenced cDNAs | FLJ-PJ | 30,063 |
| SV-PJ | 5,020 | |||
| ESTs | 5'-EST | 1,323,199 | ||
| 3'-EST | 107,239 | |||
| Other Public Database | Entirely sequenced cDNAs | 49,650 | RefSeq and Ensembl | 63,342 | ESTs | 3,862,807 |
| Gateway Entry Clone | N-Type | 17,802 | ||
| F-Type | 25,447 | |||
| Protein Analysis Data | SDS-PAGE pictures of FLJ cDNAs | 17,821 | ||
| Subcellular localization pictures of FLJ cDNAs | 10,917 | |||
FLJ cDNAs by oligo - Capping Method
- FLJ cDNA sequences of our full-length cDNAs constructed using the oligo-capping method (20)
- FLJ Human cDNA Database
To Begin Search
To begin the search, you can select the search mode from "ID Search", "Keyword Search" and "Advanced Search" which includes "BLAST Search" and "Category Search". If you want to search by an ID number (in a definitive or degenerated form) such as DDBJ/EMBL/GenBank accession number, Ensembl transcript number, Gene Symbol, FLJ ID or Sequence ID, please select the "ID Search" (Fig. 1). If you want to search by keyword(s) such as gene or protein name, any word included in gene or protein description and Gene Ontology term, please select the "Keyword Search" (Fig. 2). If you want to search by nucleotide or amino acid sequence(s), please select the "BLAST Search" (Fig. 5) included in the "Advanced Search" (Figs. 3,4). If you want to search by specific category(ies), please select the "Category Search" (Fig. 6) included in the "Advanced Search" (Figs. 3,4).
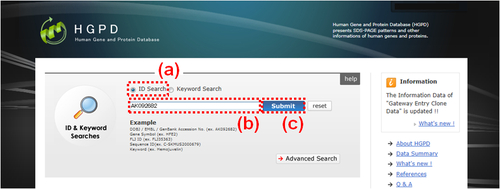
Fig. 1: HGPD top page in the case of "ID Search". (a) Please select the radio button "ID Search". (b) Please enter a term in the text box. As an example, a DDBJ/EMBL/GenBank accession number AK092682 is entered. (c) Please click the "Submit" button to start the search. If there are search hits, an "Information Overview" window (Fig. 10) is displayed.
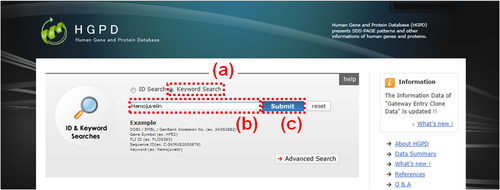
Fig. 2: HGPD top page in the case of "Keyword Search". (a) Please select the radio button "Keyword Search". (b) Please enter term(s) in the text box. As an example, a keyword Hemojuvelin is entered. (c) Please click the "Submit" button to start the search. If there are search hits, an "Keyword Search Result" window (Fig. 7) is displayed.
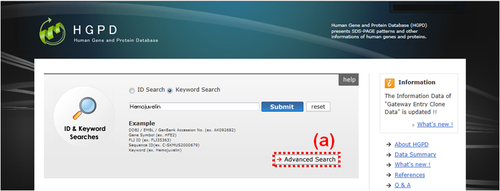
Fig. 3: HGPD top page in the case of "Advanced Search". (a) Please click the "Advanved Search" button to open an "Advanced Search" window (Fig. 4)
Advanced Search
Now, in "Advenced Search", "BLAST Search" (Fig. 5) and "Category Search" (Fig. 6) are available. In the future, other search modes will be prepared in turn.

Fig. 4: Advanced Search. (a) You can return to the top page by clicking the "HGPD" logo image. (b) If you want to use "BLAST Search", please click the tag of "BLAST Search". (c) If you want to use "Category Search", please click the tag of "Category Search". Default search mode is "BLAST Search". (d) If you want to use "ID Search" or "Keyword Search", plese click the "ID & Keyword Search" button. By doing so, you can returen to the top page (Fig. 1).
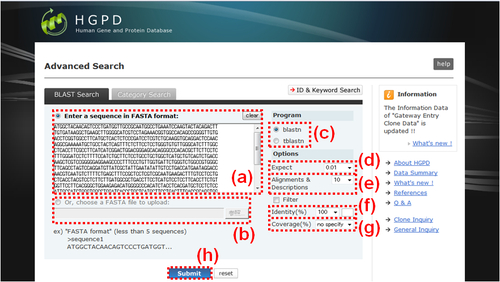
Fig. 5: BLAST Search. (a) Please input your query sequence(s) of FASTA format to the text box directly. At the first line, a name begginning at ">" can be attached. Attaching a sequence name is optional. And your multiple query sequences are available. For multiple sequences, sequence names to distinguish each sequence are indispensable. At the first line of each your sequence, each name beginning at ">" should be placed. (b) If you want to upload your query sequence(s) of FASTA format, please select the radio button here and then click "Browse.." button. In advance, please prepare the single FASTA format file as mentioned above. (c) Please select one of the search program, "blastn" or "tblastn". Default setting is "blastn". (d) Please select one of the expected values, "0.0001", "0.001", "0.01", "0.1", "1" or "10". Default value is "0.01". (e) Please select one of the number of alignments and descriptions in the BLAST output, "1", "10", "20", "50" or "100". Default value is "10". (f) Please select one of the percentage identity cutoff value, "100", "99", "98", "95" from the list box. If you select "user" from the list box, you can put any integral number (from 0 through 99) to the right side text box. Default value is "100". Please select one of the percentage coverage cutoff value, "no specify", "100", "95", "90", "80", "70", "50" or "30". Default value is "no specify". The percentage coverage is calculated by "(query sequence length) / (subject sequence length) * 100". (h) After all settings done, please click the "Submit" button to start the search". If there are search hits, a "Blast Search Results" window (Fig. 8) is newly opened.
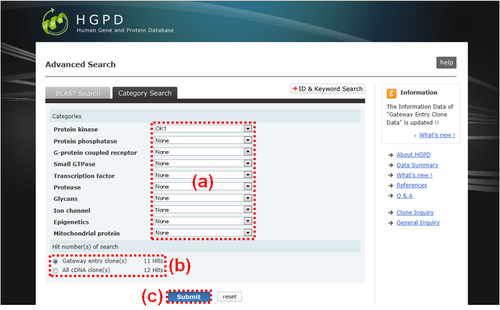
Fig. 6: Category Search. There are 10 independent first-level (parent) categories as follows: "Protein kinase", "Protein phosphatase", "G-protein coupled receptor", "Small GTPase", "Transcription factor", "Protease", "Glycans", "Ion channel", "Epigenetics" and "Mitochondrial protein". Under each parent category, there are some second-level (sub) categories. (a) Please select sub category from the list box of each category. Each default sub category is "None". As an example, "CK1" sub category of "Protein kinase" parent category is selected. (b) After selecting sub cetegory(ies), the hit number of the search are calculated and displayed. If multiple sub categories are selected, the overlap number between or among the selected sub categories are calculated. "All cDNA clone(s)" counts the number of all the hit genes, whereas "Gateway entry clone(s)" counts the number of hit genes which include Gateway entry clones. (c) After selecting the sub categoy(ies), please click the "Submit" button to display the result table "Category Search Results" (Fig. 9).
Keyword Search Results
As a result of the "Keyword Search", a "Keyword Search Results" window (Fig. 7) is displayed.
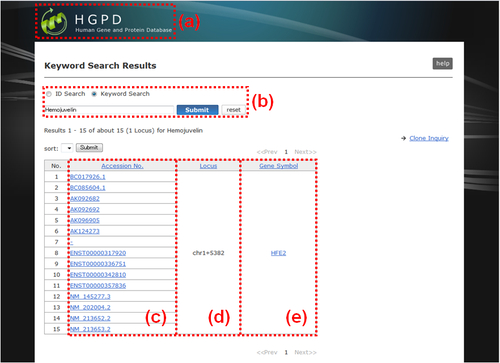
Fig. 7: Keyword Search Results. (a) You can return to the top page by clicking the "HGPD" logo image. (b) You can submit another "ID Search" or "Keyword Search" without returning to the top page in the same way mentiond above (Figs. 1,2). (c) Accession No.: The accession number of each search hit registered in the public database such as DDBJ/EMBL/GenBank and Ensembl. Each one is linked to the "Information Overview" (Fig. 10). (d) Locus: The identifier of cDNA cluster to which each search hit belongs. Each cluster is mapped on human genome by genome locus. (e) Gene Symbol: The official symbol of the gene to which each search hit belongs. Each one is linked to the NCBI's Entrez Gene page.
BLAST Search Results
As a result of the "BLAST Search", a "BLAST Search Results" window (Fig. 8) is displayed.

Fig. 8: BLAST Search Results. (a) You can return to the top page by clicking the "HGPD" logo image. (b) The identifier of each BLAST hit is displayed. Each one is linked to the "Information Overview" (Fig. 10). (c) The score for each BLAST hit is displayed. Each one is linked to the actual alignment. (d) The actual alignment of each BLAST hit is displayed. Each description line is linked to the "Information Overview" (Fig. 10).
Category Search Results
As a result of the "Category Search", a "Category Search Results" view (Fig. 9) is displayed below the category selection section (Fig. 6).
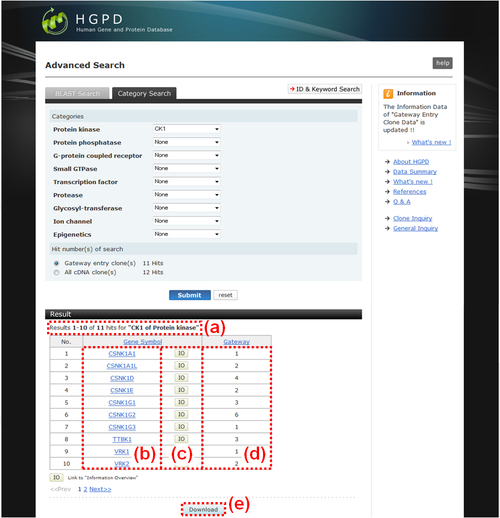
Fig. 9: Category Search Results. (a) The selected category(ies) and the numbers listed in the table below are displayed. (b) The official symbol for each hit gene. Each one is linked to the NCBI's Entrez Gene page. (c) Each "IO" button is linked to the "Information Overview" (Fig. 10). (d) The number of Gateway entry clones inclueded in each gene is displayed. (e) If you want to obtain the list of search result as a text file, please click the "Download" button.
Information Overview
A summary is presented all of information on the cluster to which the queried cDNA clone is belongs in an "Information Overview" window (Fig. 10). All data related to all cDNA members clustered in the query are shown. Almost all links used in HGPD are connected through this page.
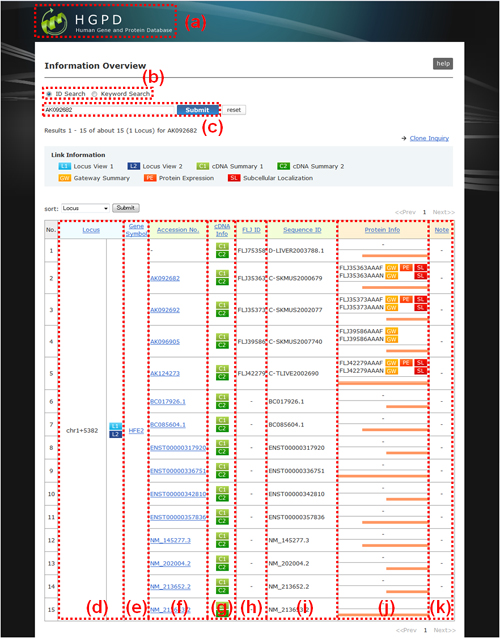
Fig. 10: Information Overview. (a) The user can return to the top page by pressing the "HGPD" logo. (b) Another search can be started by selecting ID or Keyword on the menu, and (c) entering the ID or keyword into the textbox above without returning to the top page. (d) Locus: A serial cluster number is presented. For details, see Genome mapping and clustering (13, 14). Locus number "chr1+5382" is presented as an example, where chr1, + and 5382 mean the human chromosome number, specification of the strand (plus (5'->3' strand in the direction of p arm to q arm) or minus (5'->3' strand in the direction of q arm to p arm)) on which clusters have been assembled, and the cluster number as counted consecutively from the 5' end of each strand of chr.1, respectively. L1 and L2 are linked to "Locus View 1" and "Locus View 2 - one direction", respectively. (e) Gene Symbol: The Gene Symbol appeared in the Entrez Gene database. (f) Accession No. The registered ID in the public database, such as DDBJ/EMBL/GenBank accession number or Ensembl transcript number, is described for each cDNA clone. As the newest version at the freezing date is usually adopted, no version number is displayed. (g) cDNA Info. Buttons "C1" and "C2" are linked to "C1: cDNA Summary 1" and "C2: cDNA Summary 2", respectively. Information on cDNA clones including sequences and homology search results is presented on these pages. (h) FLJ ID. The FLJ ID number of the FLJ cDNAs is shown. Any cDNA clone that has not been assigned an FLJ number is designated as "-". FLJ clones were consequently turned out to have 3 kinds of IDs: "DDBJ/EMBL/GenBank Accession No."; "FLJ ID", and "Primary Clone ID" (See relation table). (i) Sequence ID. For an authentic FLJ cDNA clone that holds a primary clone ID, the ID is designated with the prefix "C-" (cf. "F-" and "R-", showing 5'- and 3'-onepass sequences of FLJ cDNAs, respectively). For cDNA sequences other than FLJ sequences, an accession number for DDBJ/EMBL/GenBank is provided. (j) Protein Info. Several types of data can be retrieved through this window, including Gateway entry clones and protein expression. And the inclusion region of open reading frame (ORF) is easily indicated in a red line. It is note that if some locus are hit in "Information Overview", the position of N-/C- terminal of ORF region of each locus is actually different. (k) Note. A note of "Protein Info" is shown. See the detail of "Note".
Linkage of Windows
As to linkage of windows of HGPD, some buttons open new windows and other buttons display a picture in the current window. Windows for C1, C2, GW, and PE which show various pictures focusing on the same cDNA clone open in the same window, as relationship is essentially one vs. one. Other windows such as "Information Overview", L1, L2, and L2' that describe multiple cDNA clones or clusters will open a new window when transferred, as translocation is usually one vs. multiple (irreversible).
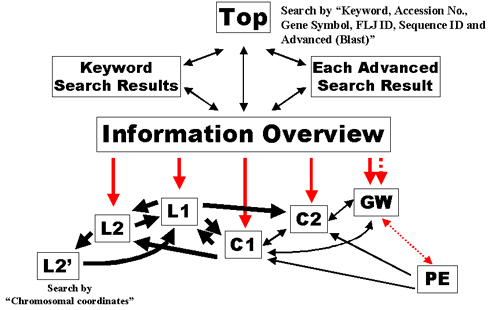
Fig. 11: Linkage of windows. "Opening a new window" and loading in the current window are indicated by thick and thin arrows, respectively. The flow from the "Information Overview" window, which is the central hub in HGPD displayed in Figure 2, is indicated by red arrows. PE is linked with the "Information Overview" page through a button in GW, which is indicated by dashed arrows. Movement within C1, C2, GW and PE will replace the previous page without opening a new window, preventing increase of the number of opened windows.
L1: Locus View 1
The genome mapping results for a queried locus are illustrated.
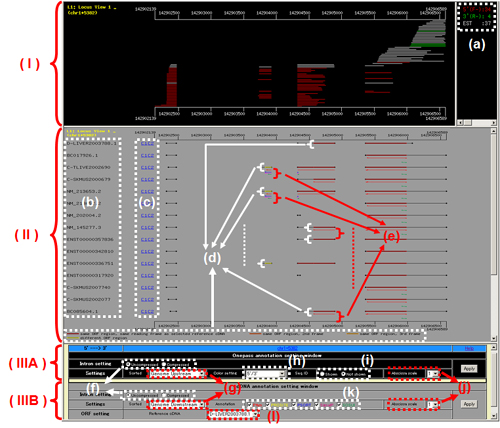
Fig. 12: L1: Locus View 1. Genome mapping of (I) ESTs and (II) Full-length cDNAs without ESTs. Setting buttons are located in the lower part (IIIA and IIIB). EST (I and IIIA) and full-length (II and IIIB) windows including setting ones are displayed in the black and gray-background, respectively. (a) 5'-ESTs of FLJ cDNAs, 3'-ESTs of FLJ cDNAs and other public ESTs are shown in red, green and white, respectively. These colors can be changed by the button to "FLJ OnePass: red, public ESTs: white" shown in (h). (b) Sequence ID. (c) C1 and C2 are linked with "C1: cDNA Summary 1" window and "C2: cDNA Summary 2" window, respectively. (d) ORF information. ORF regions are designated in relation to the frame of a selected reference ORF. (e) Regions of domains and motifs predicted by Pfam, PROSITE, PSORT, SignalP and SOSUI can be presented. Detailed data are presented in the lower part of "C2: cDNA Summary 2" (Fig. 10B). (f) Intron setting button. As intronic regions only can be removed using the "compressed" setting, splicing-variant cDNAs or unspliced intron regions can be well analyzed. (g) Sorting of cDNAs can be performed according to several conditions. Genome Upstream: Sequences that hold further upstream regions are piled up from the bottom; Genome Downstream: Sequences that hold further downstream regions are piled up from the bottom. (h) Color setting of ESTs. 5'/3': 5'-ESTs(FLJ), red; 3'-ESTs(FLJ), green; public ESTs, white. OnePass/EST: FLJ OnePass, red; public ESTs, white. (i) Sequence ID of ESTs can be shown, although space between each EST at region (I) is expanded. (j) Abscissa scale. Currently only available only 1. (k) Setting buttons for Pfam, PROSITE, PSORT, SignalP and SOSUI. (l) Reference cDNA that determines the standard ORF can be selected.
L2: Locus View 2 - one direction
A bird's eye view of clusters is presented. Cluster chr1+5382 harboring a queried sequence is highlighted by a green line. Other clusters are shown in red lines. In each cluster, the 5'- and 3'- most distal ends are tied by a line with arrows at both boundaries.
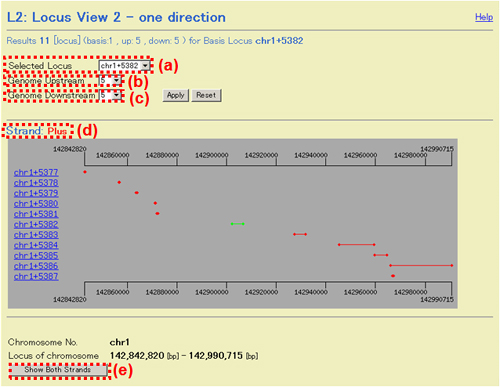
Fig. 13: L2: Locus View 2 - one direction. (a) Selection of centered locus. Locus is ( as default) specified by centering on the entered ID. Setting the number of (b) upstream or (c) downstream side loci. (d) Polarity of the strand. The 5'- to 3'- strand in the direction of p arm to q arm is defined as "Plus". Cluster number (chr1+5382 in this case) including the selected cDNA (sequence ID) is represented by a green line. The 5'- and 3'-terminal ends of this cluster are tied with a line. (e) For displaying the clusters of both strands (see L2').
L2: Locus View 2 - both direction
Clicking a "Show Both Strands" button in an L2 picture (Fig. 13) opens an "L2': Locus View 2 - both directions" window. Although the boundary of L2' picture is the same as L1, all the clusters whose any part is included are displayed. Once an "L2'- both directions" window opens, one can open any window by specifying a chromosome number and nucleotide positions (current maximum number of clusters which can be open for both strands is 50, respectively). Therefore it would be said that L2' has some search function (Fig. 11).
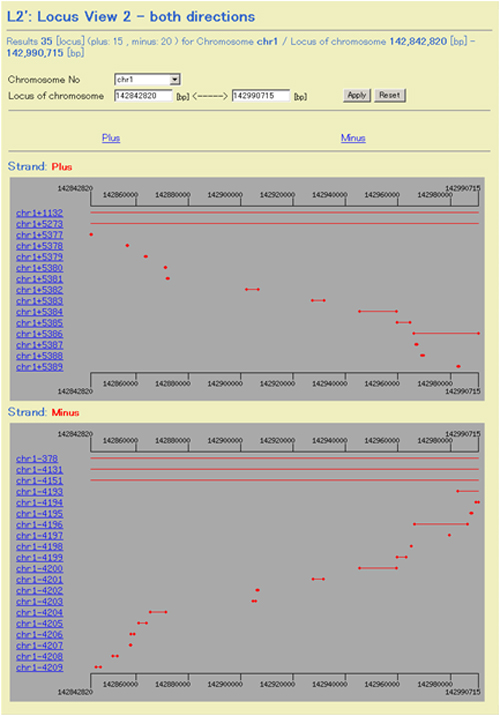
Fig. 14: L2: Locus View 2 - both direction. Regions of clusters mapped on both strands are shown by red lines. Cluster numbers for plus and minus are counted from the distal side of the p arm and q arm, respectively. An identical or similar number of clusters on the plus and minus strands therefore does not indicate proximity of these clusters.
C1: cDNA Summary 1
Sequence ID, locus ID, numbers and kinds of cDNA sequences included in a cluster and alignment data of the selected cDNA clone are presented.
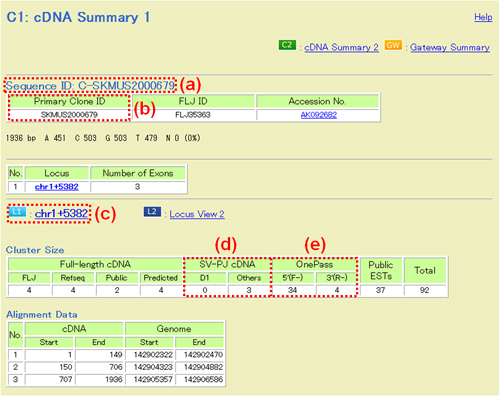
Fig. 15: C1: cDNA Summary 1. (a) Sequence ID. (b) Primary Clone ID: The initial 3-5 characters stand for names of cells or tissues from which mRNAs were purified (For details, see a complete list). (c) Locus ID. (d) SV-PJ cDNA. SV-PJ stands for Splicing Variant Project, one of the sub-projects of the NEDO "Functional analysis of human proteins" project. (e) OnePass: The number of FLJ ESTs. 5'(F-), 5'-ESTs; 3'(R-), 3'-ESTs.
C2: cDNA Summary 2
In the upper part of "cDNA Summary 2", amino acid and nucleotide sequences are represented. Red characters in nucleotide sequences indicate the ORF region.
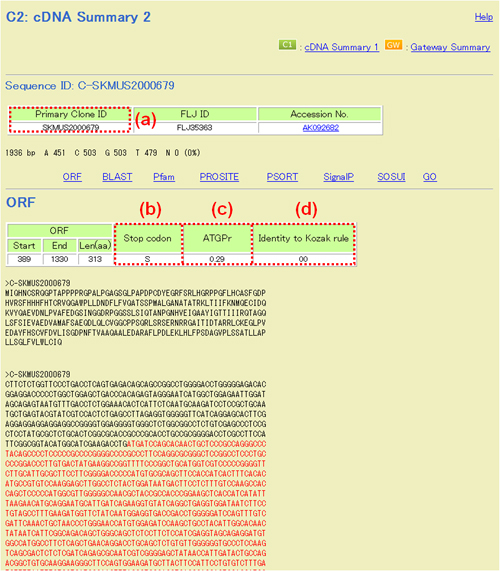
Fig. 16A: Upper part of "C2: cDNA Summary 2". (a) Primary Clone ID. See Fig. 15(b). (b) Stop codon: S and L stand for presence and absence of Stop codon, respectively. (c) and (d) Refer to reference 21.

Fig. 16B: Lower part of "C2: cDNA Summary 2". Functional motifs and domains, leader sequences and transmembrane domains were inferred using BLAST, Pfam, PROSITE, PSORT, SignalP, SOSUI and GO.
GW: Gateway Summary
The Gateway cloning system (Invitrogen, CA, USA) is based on versatile expression vectors (11). We have therefore adopted this system and constructed human Gateway entry clones named as Human Proteome Expression Resource (HUPEX) from full-length cDNAs. Detail informations described in our paper in Nature methods, 2008 (12).
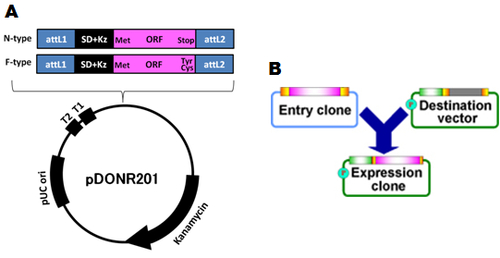
Fig. 17: (A) The structure of human Gateway entry clones. The base plasmid is pDONR201 (Invitrogen) and the attL1-sites (blue), SD-Kz (black) and ORF (pink) is cloned. There are two ORF-types, one is N-type harboring stop codon at the end of ORF, another is F-type without stop codon (replaced to Tyr or Cys). attL1 and attL2, Gateway recombination sites; SD, Shine-Dalgano sequence; Kz, Kozak sequence; ORF, open reading frame; T1 and T2, transcriptional terminators; Kanamycin, kanamycin resistant gene. (B) Gateway system. An expression clone to be utilized for expression is produced by recombination of an entry clone and a destination vector.
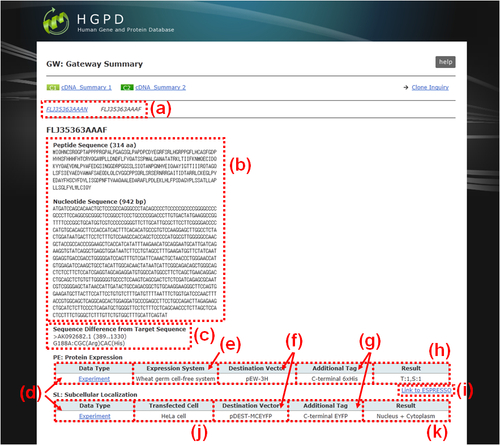
Fig. 18: GW: "Gateway Summary". Sequence information of the Gateway entry clone and summary of experimental or prediction results obtained by using the clone are presented. (a) Button for switching to the another ORF-type clone. (b) Amino acid and nucleotide sequences of human ORF regions of the clone. (c) Deviations from the reference sequence. (d) Link button to the window to view detailes of experimental ('Experiment' button) or prediction ('Prediction' button) results. (e) Protein expression system used in the experiment. (f) Destination vector used for construction of the expression clone. (g) Fused tag with the recombinant protein and the location. (h) Result of SDS-PAGE analysis for protein expression and solubility. T, total fraction; S, supernatant fraction; 1, detected; 0, not-detected. (i) Link button to the submission window of ESPRESSO which is a sequence-based predictor for estimating protein expression and solubility. (j) Host cell used for the subcellular localization analysis. (k) Intracellular compartment(s) of the recombinant protein. There are 12 major compartmentation categories as follows: 'Cytoplasm', 'Nucleus', 'Cytoplasm + Nucleus', 'Endoplasmic reticulum', 'Golgi apparatus', 'Endoplasmic reticulum + Golgi apparatus', 'Plasma membrane', 'Endoplasmic reticulum + Golgi apparatus + Plasma membrane', 'Cytoplasmic vesicles', 'Mitochondria', 'Peroxisome', 'Cytoskeleton'. Some proteins which could not be classified into these categories were assigned into 'Others'. In the cases that the proteins belonged to more than two categories in a single cell and different categories between independent cells, they were assigned 'Bilocation' and 'Mixed', respectively.
PE: Protein Expression
The SDS-PAGE patterns of expressed proteins are displayed.
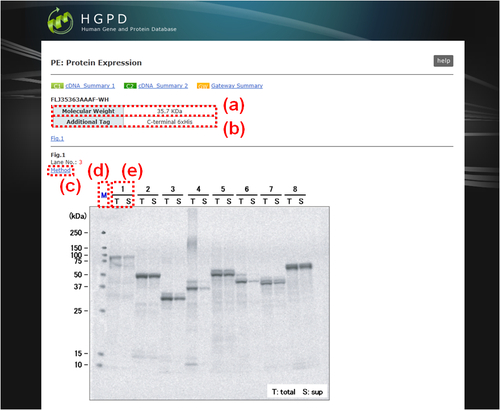
Fig. 19: "PE: Protein Expression". (a) Theoretical molecular weight of the tagged recombinant protein. (b) Fused tag with the recombinant protein and the location. (c) Link button to the window to indicate the protocol for this experiment. (d) M, Marker. (e) Number, lane number in this SDS-PAGE analysis; T, total fraction; S, supernatant fraction. Selected protein is represented in the column "Lane No." by a red character.
SL: Subcellular Localization
The subcellular localization pictures of expressed proteins are displayed.
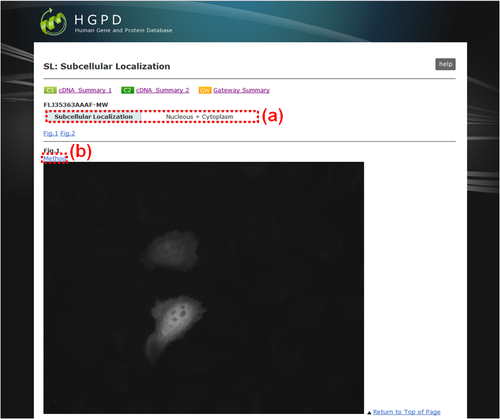
Fig. 20: "SL: Subcellular Localization". (a) Intracellular compartment(s) of the recombinant protein. There are 12 major compartmentation categories as follows: 'Cytoplasm', 'Nucleus', 'Cytoplasm + Nucleus', 'Endoplasmic reticulum', 'Golgi apparatus', 'Endoplasmic reticulum + Golgi apparatus', 'Plasma membrane', 'Endoplasmic reticulum + Golgi apparatus + Plasma membrane', 'Cytoplasmic vesicles', 'Mitochondria', 'Peroxisome', 'Cytoskeleton'. Some proteins which could not be classified into these categories were assigned into 'Others'. In the cases that the proteins belonged to more than two categories in a single cell and different categories between independent cells, they were assigned 'Bilocation' and 'Mixed', respectively. (b) Link button to the window to indicate the protocol for this experiment.
Note of Protein Info
In genome mapping for each cDNA sequence, some cDNA sequences are mapped more than two locus on human genome sequences. In this case, we take notice of ORF region (Gatway entry clone) only as "Note of Protein Info". In HGPD, there are two of major mapping patterns, and each pattern is named "Chimeric", "Duplicated" and "Unmapping", respectively. And "C", "D" and "*" are displayed at the cloumn "Note" of "Information Overview", respectively (Fig. 21, 22 and 23). In "Note" of "Protein Info", although each ORF region mapped on human genome suquence is evaluated now, other evaluation about "Protein Info" will be performed in the future.
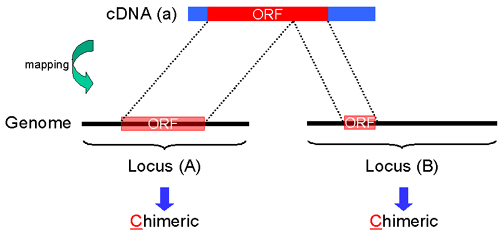
Fig. 21: "C". A part of ORF region of cDNA sequence(a) is mapped on human gene sequence (Locus (A)). And the other part of ORF region of cDNA sequence(a) is mapped on human genome sequence (Locus (B)). In this case, both "Protein Info" of Gateway entry clone in Locus (A) and Locus (B) are chimeric, and "C" is displayed at the both columns of "Note".
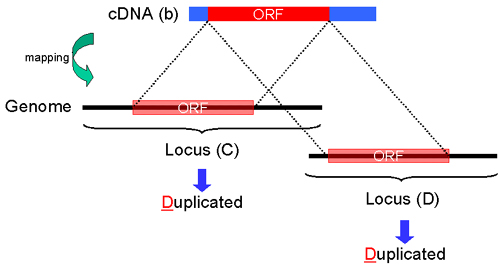
Fig. 22: "D". An ORF region of cDNA sequence(b) is mapped on each of different human genome suquence (Locus (C) and Locus (D)). In this case, both "Protein Info" of Gateaway entry clone in Locus (C) and Locus (D) are duplicated, and "D" is displayed at the both columns of "Note".
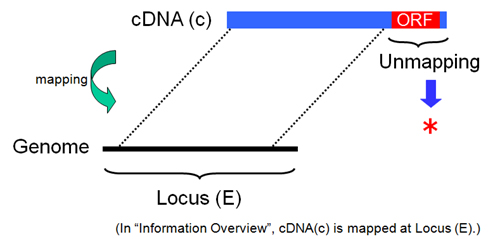
Fig. 23: "*". An ORF region of cDNA sequence(c) is unmapped on human genome suquence. In this case, "Protein Info" of Gateaway entry clone in Locus (E) is not detected, and "*" is displayed at the column of "Note". Some ORF region of cDNA sequence(c), however, is mapped on human genome sequence at another locus (not Locus(E)). About this, HGPD has no information. Please confirm it by blast search and so on.
Q & A
Under Construction
Clone Inquiry
Please see the Clone Inquiry page.
Questions & Comments
If you have any questions, please feel free to contact JBIC
Future Tasks
We hope to develop the following matters.
- Classification of the Gateway entry clones under protein function, continuously
- Correspondence accession number of cDNA clones and gene symbol to protein accession number (e.g. NP and XP number of NCBI reference sequence, entry name and accession of Swiss-Prot and so on)
- Improvement of the method of the "ID Search" tool with protein accession number
References
- International Human Genome Sequencing Consortium. (2004) Finishing the euchromatic sequence of the human genome. Nature 431, 931-945.
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nature Genet. 36, 40-45.
- Kimura, K., Wakamatsu, A., Suzuki, Y., Ota, T., Nishikawa, T., Yamashita, R., Yamamoto, J., Sekine, M., Tsuritani, K., Wakaguri, H., et al. (2006) Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16, 55-65.
- Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, O. K., Barrero, A. R., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. (2004). Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biology 2, 0001-0020.
- Nomura, N., Miyajima, N., Sazuka, T., Tanaka, A., Kawarabayasi, Y., Sato, S., Nagase, T., Seki, N., Ishikawa, K., and Tabata, S. (1994) Prediction of the Coding Sequences of Unidentified Human Genes. I. The Coding Sequences of 40 New Genes (KIAA0001-KIAA0040) Deduced by Analysis of Randomly Sampled cDNA Clones from Human Immature Myeloid Cell Line KG-1. DNA Res. 1, 27-35.
- Ohara, O., Nagase, T., Ishikawa, K., Nakajima, D., Ohira, M., Seki, N., and Nomura, N. (1997). Construction and characterization of human brain cDNA libraries suitable for analysis of clones encoding relatively large proteins. DNA Research 4, 53-59.
- Mammalian Gene Collection Program Team. (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 99, 16899-16903.
- Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Bocher, M., Blocker, H., Bauersachs, S., Blum, H., et al. (2001) Toward a Catalog of Human Genes and Proteins: Sequencing and Analysis of 500 Novel Complete Protein Coding Human cDNAs. Genome Res. 11, 422-435.
- Hu, R., Han, Z., Song, H., Peng, Y., Huang, Q., Ren, S., Gu, Y., Huang, C., Li, Y., Jiang, C., et al. (2000) Gene expression profiling in the human hypothalamus-pituitary-adrenal axis and full-length cDNA cloning. Proc. Natl. Acad. Sci. USA 97, 9543-9548.
- Temple, G., Lamesch, P., Milstein, S., Hill, DE., Wagner, L., Moore, T. and Vidal, M. (2006) proteome: developing expression clone resources for the human genome. Hum Mol Genet. 15, R31-43.
- Hartley, J., Temple, G. and Brasch, M. (2000) DNA Cloning Using In Vitro Site-Specific Recombination. Genome Res. 10, 1788-7895.
- Goshima, N., Kawamura, Y., Fukumoto, A., Miura, A., Honma, R., Sato, R., Wakamatsu, A., Yamamoto, J., Kimura, K., Nishikawa, T., et al. (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nature Methods 5, 1011-1017.
- Otsuki, T., Ota, T., Nishikawa, T., Hayashi, K., Suzuki, Y., Yamamoto, J., Wakamatsu, A., Kimura, K., Sakamoto, K., Hatano, N., et al. (2005) Signal sequence and keyword trap in silico for selection of full-length human cDNAs encoding secretion or membrane proteins from oligo-capped cDNA libraries. DNA Res. 12, 117-126.
- Nishikawa, T., Ota, T. and Isogai, T. (2000) Prediction whether a human cDNA sequence contains initiation codon by combining statistical information and similarity with protein sequences. Bioinformatics 16, 960-967.
- Madin, K., Sawasaki, T., Ogasawara, T. and Endo, Y. (2000) A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc Natl. Acad. Sci. USA 97, 559-64.
- Sawasaki, T., Ogasawara, T., Morishita, R. and Endo, Y. (2002) A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA 99, 14652-14657.
- Kisu, Y., Togashi, T., Sono, S., Mochizuki, H., Yamaguchi, K., Takada, N., Matsukura, S., Seki, T., Kuwayama, H., Yoshida, A., et al. Atlas of Subcellular Localization of 17,000 Human Proteins. (in preparation).
- Maruyama, Y., Wakamatsu, A., Kawamura, Y., Kimura, K., Yamamoto, J., Nishikawa, T., Sugano, S., Goshima, N., Isogai, T. and Nomura, N. (2009) Human Gene and Protein Database (HGPD): a novel database presenting a large quantity of experiment-based results in human proteomics. Nucleic Acid Res. 37, D762-766.
- Wakamatsu, A., Yamamoto, J., Kimura, K., Nishikawa, T., Sugano, S., Nomura, N., and Isogai, T. (2007). FLJ human cDNA database focused on mRNA variation. The Proceedings of the 2007 Annual Conference of Japanese Society for Bioinformatics (JSBi2007), P048-1 - P048-2.
- Suzuki, Y., Yoshitomo-Nakagawa, K., Maruyama, K., Suyama, A. and Sugano, S. (1997) Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library. Gene 200, 149-56.
- Salamov, A. A., Nishikawa, T. and Swindells, M. B. (1998) Assessing protein coding region integrity in cDNA sequencing project. Bioinformatics 14, 384-390.
Links
- FLJ Human cDNA Database
- H-Inv DB
- ESPRESSO
- FLJ-DB
- DDBJ
- NEDO Full-length human cDNA sequencing Project
- HUGE
- NCBI
- MGC
- The German Human cDNA Project
- UniGene
- CGAP
- SAGE
- TIGR Gene Indices
- STACK
- BODYMAP
- NITE Biological Resource Center
- AIST Biomedicinal Information Research Center
- Japan Biological Informatics Consortium
- Research Association for Biotechnology
- Reverse Proteomics Research Institute
- About HGPD
- What's New !
- Data Summary
- Oligo-Capping Method
- To Begin Search
- Advenced Search
- Keyword Search Results
- BLAST Search Results
- Category Search Results
- Information Overview
- Linkage of windows
- Locus View 1
- Locus View 2
- Locus View 2'
- cDNA Summary 1
- cDNA Summary 2
- Gateway Summary
- Protein Expression
- Subcellular Localization
- Note of Protein Info
- Q & A
- Clone Inquiry
- Questions & Comments
- Future Tasks
- References
- Links

