Method of Subcellular Localization Analysis
|
Method of Subcellular Localization Analysis
|
For analysis of a large number of proteins, we have developed a system that comprises an automatic plate-handling robot, a cell-imaging apparatus using an optical microscope, and an image archive server. HeLa cells were transfected with expression clones and fixed after 24 hours. Cells were visualized by fluorescence, and microscopic images of cells were obtained using an automatic microscope system and/or semi-automatic microscope system. Image data (each image is about 1 MB) were archived in the data management system on the server through the local network. Subcellular localization patterns for cell images were annotated manually (Fig. A).
Fig. A: Flow of subcellular localization analysis.
High-throughput preparation of expression clones (Gateway system) 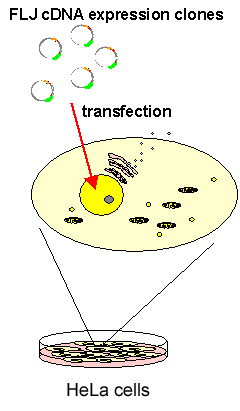
↓ Cell Plating ↓ plate HeLa cells in 96 well glass plate incubate at 37°C for 24 h Transfection ↓ transfect 100ng of expression clone DNA with TransIT solution (Mirus) incubate at 37°C for 24 h Fixaction ↓ fix cells with 4% formaldehyde solution incubate at room temperature for 1 h Microscopic imaging
(automatic system, semi-automatic system)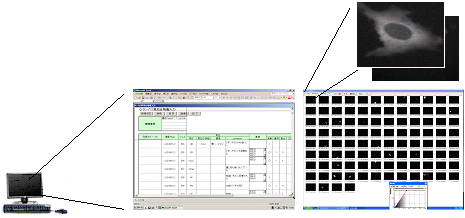
↓ Judgment of localization
(manually)
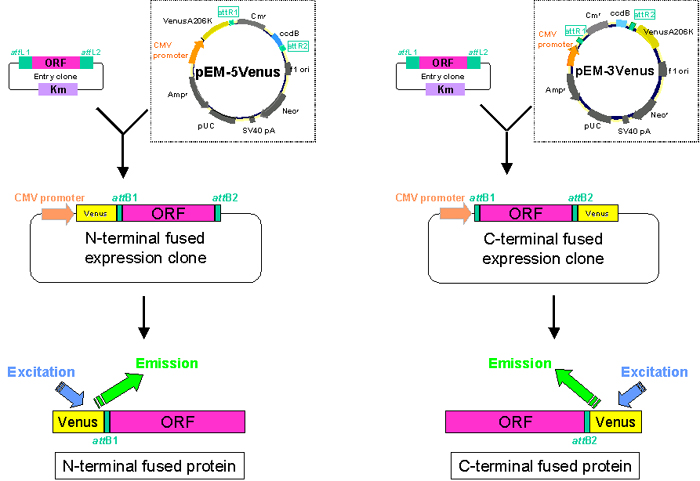
For visualizing techniques with fluorescence, Gateway destination vectors harboring a modified-YFP protein gene, VenusA206K, were constructed. Over 30,000 FLJ cDNA expression clones (N-terminal tagged type and C-terminal tagged type) were prepared using this vector (Fig. B).
Fig. B: Destination vectors for localization analysis. ORF regions of human genes have been cloned into destination vectors pMW-VN or pMW-VC which can fuse human ORFs with Venus protein either at the N- or C-terminial end.
 |